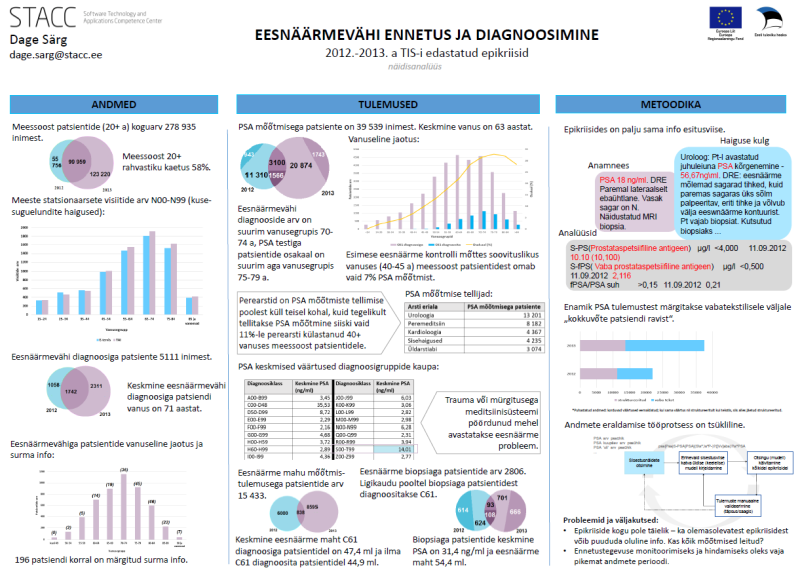
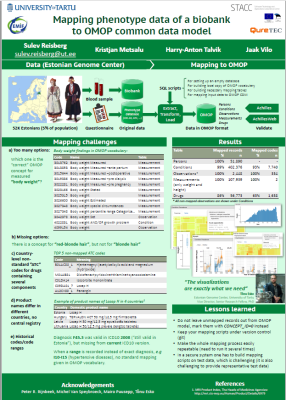
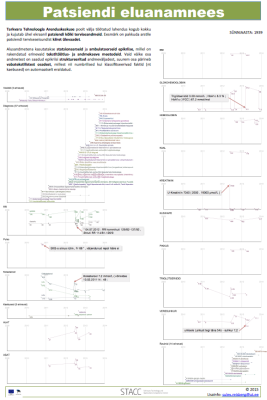
Below is a list of our articles published in scientific journals together with a selection of posters presented in various international conferences. View also the list of completed bachelor, masters and doctoral thesis.
Haug M, Oja M, Pajusalu M, Mooses K, Reisberg S, Vilo J, Giménez AF, Falconer T, Danilović A, Maljkovic F, Dawoud D, Kolde R.
Markov modeling for cost-effectiveness using federated health data network
Journal of the American Medical Informatics Association, 2024; ocae044, doi: 10.1093/jamia/ocae044
Markus AF, Rijnbeek PR, Kors JA Burn E, Duarte-Salles T, Haug M, Kim C, Kolde R, Lee Y, Park HS, Park RW, Prieto-Alhambra D, Reyes C, Krishnan JA, Brusselle GG, Verhamme KMC
Real-world treatment trajectories of adults with newly diagnosed asthma or COPD
Mercadé-Besora N, Li X, Kolde R, Trinh NTH, Sanchez-Santos MT, Man WY, Roel E, Reyes C, Delmestri A, Nordeng HME, Uusküla A, Duarte-Salles T, Prats C, Prieto- Alhambra D, Jödicke AM, Català M
Heart 2024;0:1–9. doi:10.1136/heartjnl-2023-323483
Oja M, Tamm S, Mooses K, Pajusalu M, Talvik HA, Ott A, Laht M, Malk M, Lõo M, Holm J, Haug M, Šuvalov H, Särg D, Vilo J, Laur S, Kolde R, Reisberg S
JAMIA Open, 2023: 6 (4). DOI: 10.1093/jamiaopen/ooad100
Uusküla, A; Oja, M; Tamm, S; Tisler, A; Laanpere, M; Padrik, L; Nygard, M; Reisberg, S; Vilo, J; Kolde, R.
JAMA Network Open, 2023:6 (2), e2254075. DOI: 10.1001/jamanetworkopen.2022.54075
Mooses K; Vesilind, K; Oja M; Tamm S; Haug M; Kalda R; Suija K, Tisler A, Meister T; Malk M, Uusküla A; Kolde R.
The use of prescription drugs and health care services during the 6‑month post‑COVID‑19 period
Scientific Reports, 2023:13, 11638100394. DOI: 10.1038/s41598-023-38691-9.
Gandaglia G, Pellegrino F, Golozar A, De Meulder B, Abbott T, Achtman A, Omar MI, Alshammari T, Areia C, Asiimwe A, Beyer K, Bjartell A, Campi R, Cornford P, Falconer T, Feng Q, Gong M, Herrera R, Hughes N, Hulsen T, Kinnaird A, Lai LYH , Maresca G, Mottet N, Oja M, Prinsen P, Reich C, Remmers S, Roobol MJ, Sakalis V, Seager S, Smith EJ, Snijder R, Steinbeisser C, Thurin NH, Hijazy A, van Bochove K, Van den Bergh RCN, Van Hemelrijck M, Willemse PP, Williams AE, Kermani NZ, Evans-Axelsson S, Briganti A, N’Dow J, on behalf of the PIONEER Consortium
European Urology, 2023. DOI: 10.1016/j.eururo.2023.06.012
Voss EA, Shoaibi A, Lai LYH, Blacketer C, Alshammari T, Makadia R, Haynes K, Sena AG, Rao G, van Sandijk S, Fraboulet C, Boyer L, Le Carrour T, Horban S, Morales DR, Roldán JM, Ramírez-Anguita JM, Mayer A, de Wilde M, John LH, Duarte-Salles T, Roel E, Pistillo A, Kolde R, Maljkovi F, Denaxas S, Papez V, Kahn MG, Natarajan K, Reich C, Secora A, Minty EP, Shah NH, Posada JD, Morales MTG, Bosca D, Juanino HC, Holgado AD, Jiménez MP, Balazote PS, Barrio NG, Sen S, Üresin AY, Erdogan B, Belmans L, Byttebier G, Malbrain MLNG, Dedman DJ, Cuccu Z, Vashisht R, Butte AJ, Patel A, Dahm L, Hana C, Arshad FBF, Ostropolets A, Nyberg F, Hripcsak G, Suchard MA,
Prieto-Alhambra D, Rijnbeek PR, Schuemie MJ, Ryan PB
eClinicalMedicine. Part of THE LANCET Discovery Science, 2023:58. DOI: 10.1016/j.eclinm.2023.101932
Rosenberg M, Thetloff M, Tamm S, Kuusk K, Reisberg S, Vilo J
Kroonilise neeruhaiguse levimus Eesti e-tervise andmete alusel
Meister, T; Pisarev, H; Kolde, R; Kalda, R; Suija, K; Milani, L; Karo-Astover, L; Piirsoo, M; Uusküla, A.
PLoS ONE, 2022:17, e0270192. DOI: 10.1371/journal.pone.0270192
Uusküla, A; Jürgenson T; Pisarev H; Kolde, R; Meister, T; Tisler, A; Suija, K; Kalda, R; Piirsoo, M; Fischer K
Long-term mortality following COVID-19 infection: a national cohort study from Estonia
The Lancet Regional Health – Europe, 2022:100394. DOI: 10.1016/j.lanepe.2022.100394.
Yang, C; Williams, Ross D; Swerdel, JN; Almeida, JR; Brouwer, ES.; Burn, E; Carmona, L; Chatzidionysiou, K; Duarte-Salles, T; Fakhouri, W; Hottgenroth, A; Jani, M; Kolde, R; Kors, JA; Kullamaa, L; Lane, J; Marinier, K; Michel, A; Stewart, HM; Prats-Uribe, A … Rijnbeek, PR.
Seminars in Arthritis and Rheumatism, 2022:56, 152050. DOI: 10.1016/j.semarthrit.2022.152050
Künnapuu, K; Ioannou, S; Ligi, K; Kolde, R; Laur, S; Vilo, J; Rijnbeek, PR; Reisberg, S
JAMIA Open, 2022:5 (1), 1−11. DOI: 10.1093/jamiaopen/ooac021
Ostrak, A; Randmets, J; Sokk, V; Laur, S; Kamm, L.
Implementing Privacy-Preserving Genotype Analysis with Consideration for Population Stratification
Cryptography, 2021:5 (3), ARTN 21. DOI: 10.3390/cryptography5030021.
Tamm, S; Raie, E; Käär, R; Oja, M; Reisberg, S
Eesti Arst, 2020:99 (1), 6−15.
Laur, S; Orasmaa, S; Särg, D; Tammo, P
EstNLTK 1.6: Remastered Estonian NLP Pipeline. Proceedings of the 12th Language Resources and Evaluation
Conference: International Conference on Language Resources and Evaluation (LREC), Marseille, France, 2020. European Language Resources Association, 7152−7160.
Krebs, K; Bovijn, J; Zheng, N; Lepamets, M; Censin, JC; Jürgenson, T; Särg, D; Abner, E; Laisk, T; Luo, Y; Skotte, L; Geller, F; Feenstra, B; Wang, W; Auton, A; Raychaudhuri, S; Esko, T; Metspalu, A; Laur, S… Fadista, J
Genome-wide Study Identifies Association between HLA-B∗55:01 and Self-Reported Penicillin Allergy
The American Journal of Human Genetics, 2020:107 (4), 612−621. DOI: 10.1016/j.ajhg.2020.08.008.
James, G; Reisberg, S; Lepik, K; Galwey, N; Avillach, P; Kolberg, L; Mägi, R; Esko, T; Alexander, M; Waterworth, D; Loomis, AK; Vilo, J
PLoS ONE, 2019:14 (4), ARTN e0215026. DOI: 10.1371/journal.pone.0215026.
Ward, DV; Hoss, AG; Kolde, R; van Aggelen, HC; Loving, J; Smith, SA; Mack, DA; Kathirvel, R; Halperin, JA; Buell, DJ; Wong, BE; Ashworth, JL; Fortunato-Habib, MM; Xu, L; Barton, BA; Lazar, P; Carmona, JJ; Mathew, J; Salgo, IS; Gross, BD … Ellison, RT, III
Integration of genomic and clinical data augments surveillance of healthcare-acquired infections
Infection Control and Hospital Epidemiology, 2019:40 (6), 649−655. DOI: 10.1017/ice.2019.7
Liivlaid, H; Eigo, N; Reisberg, S
Eriarstiabi haigestumusstatistika võrdlus Tervise Arengu Instituudi ja Eesti Haigekassa andmetel
Eesti Arst, 2019:1, 17−26
Palover, M; Leitsalu, L; Krebs, K; Lall, K; Kals, M; Nikopensius, T; Reigo, A; Milani, L; Fischer, K; Magi, R; Allik, A; Kolk, B; Metsalu, K; Puusepp, M; Mikkel, K; Tammesoo, M; Pintsaar, E; Temberg, T; Reisberg, S; Vilo, J … Metspalu, A.
Reporting genetic risks to participants of the Estonian Biobank, Genome Center
European Journal of Human Genetics, 2019:27 (1), 544−545
Reisberg, S; Krebs, K; Lepamets, M; Kals, M; Mägi, R; Metsalu, K; Lauschke, VM; Vilo, J; Milani, L
Genetics in Medicine, 2019 DOI: 10.1038/s41436-018-0337-5
Mooses, K; Oja, M; Reisberg, S; Vilo, J; Kull, M
BMC Public Health, 2018:18. DOI: 10.1186/s12889-018-5752-7.
Bogdanov, D; Kamm, L; Laur, S; Sokk, V
IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2018:15 (5), 1427−1432. DOI: 10.1109/TCBB.2018.2858818.
Bogdanov, D; Kamm, L; Laur, S; Sokk, V
Rmind: A Tool for Cryptographically Secure Statistical Analysis
IEEE Transactions on Dependable and Secure Computing, 2018:15 (3), 481−495. DOI: 10.1109/TDSC.2016.2587623
Reisberg, S; Iljasenko, T; Läll, K; Fischer, K; Vilo, J
PLoS ONE, 2+17:12 (7), e0179238−e0179238. DOI: 10.1371/journal.pone.0179238.
Muromägi, A; Sirts, K; Laur, S
Linear Ensembles of Word Embedding Models
Proceedings of the 21st Nordic Conference on Computational Linguistics: NoDaLiDa, Gothenburg, Sweden, 22.-24.05.2017. Linköping University Electronic Press, 96−104.
Reimand J; Arak T; Adler P; Kolberg L; Reisberg S; Peterson H; Vilo J
g:Profiler-a web server for functional interpretation of gene lists (2016 update)
Nucleic Acids Research, 2016, gkw199. DOI: 10.1093/nar/gkw199
Roberto, G; Leal, I; Sattar, N; Loomis, AK; Avillach, P; Egger, P; van Wijngaarden, R; Ansell, D; Reisberg, S; Tammesoo, ML; Alavere, H; Pasqua, A; Pedersen, L; Cunningham, J; Tramontan, L; Mayer, MA; Herings, R; Coloma, P; Lapi, F; Sturkenboom, M … Gini, R.
Identifying Cases of Type 2 Diabetes in Heterogeneous Data Sources: Strategy from the EMIF Project
PLoS ONE, 2016:11 (8), e0160648. DOI: 10.1371/journal.pone.0160648.
Orasmaa, S; Petmanson, T; Tkatšenko, A; Laur, S; Kaalep, HJ
EstNLTK – NLP Toolkit for Estonian
Proceedings of the Tenth International Conference on Language Resources and Evaluation (LREC 2016): The International Conference on Language Resources and Evaluation; Portorož, Slovenia. 2460−2466.
Kolde, R; Märtens, K; Lokk, K; Laur, S; Vilo, J
Bioinformatics, 2016, btw304. DOI: 10.1093/bioinformatics/btw304.
Reisberg, S ; Talvik, HA; Koppel, K; Laur, S; Vilo, J
Bogdanov, D; Kamm, L; Laur, S; Pruulmann-Vengerfeldt, P; Talviste, R; Willemson, J
Privacy-preserving statistical data analysis on federated databases
Privacy Technologies and Policy: Annual Privacy Forum 2014, 20.-21. mai 2014, Ateena, Kreeka. DOI: 10.1007/978-3-319-06749-0_3.
Reisberg, S; Sirel, R; Kalda, R; Merzin, M; Pruulmann, J; Vilo, J
Elektrooniliste terviselugude analüüsimise võimalused Tartu perearstide infosüsteemi näitel
Eesti Arst, 2013:92 (8), 452−459.
Kamm, L; Bogdanov, D; Laur, S; Vilo, J
A new way to protect privacy in large-scale genome-wide association studies
Bioinformatics, 2013:29 (7), 886−893. DOI: 10.1093/bioinformatics/btt066.
Tkachenko, A; Petmanson, T; Laur, S
Named Entity Recognition in Estonian
Proceedings of the 4th Biennial International Workshop on Balto-Slavic Natural Language Processing, Sofia, Bulgaria, 8-9 August 2013. ACL: Omnipress, 78−83.
Bogdanov, D; Jagomägis, R; Laur, S
A universal toolkit for cryptographically secure privacy-preserving data mining
In: Proceedings of the Pacific Asia Workshop on Intelligence and Security Informatics 2012 (112−126).
Kolde, R; Laur, S; Adler, P; Vilo, J
Robust Rank Aggregation for gene list integration and meta-analysis
Bioinformatics, 2012:28 (4), 573−580. DOI: 10.1093/bioinformatics/btr709
Tretyakov, K.; Laur, S.; Vilo, J
G = MAT: Linking Transcription Factor Expression and DNA Binding Data
PLoS ONE, 2011:6 (1), e14559. DOI: 10.1371/journal.pone.0014559.
Ilmjärv, S; Luuk, H; Laur, S; Vasar, E; Vilo, J
Non-parametric probe-level estimation of differential expression
In: . 19th Annual International Conference on Intelligent Systems for Molecular Biology and 10th European Conference on Computational Biology, Vienna (Austria), 2011.
Jänes, J; Laur, S; Kolka, I
Detection and characterisation of Hα emission lines from Gaia BP/RP spectra
C.A.L. Bailer-Jones. Classification and discovery in large astronomical surveys (71−75). Melville, New York: AIP.
Bogdanov, D; Laur, S; Willemson, J
Sharemind: a framework for fast privacy-preserving computations
Computer Security – ESORICS 2008: 13th European Symposium on Research in Computer Security Málaga, Spain, 192−206. DOI: 10.1007/978-3-540-88313-5_13.
Laur, S; Lipmaa, H; Mielikäinen, T
Cryptographically Private Support Vector Machines
KDD-2006: The Twelfth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Philadelphia, USA, 2006, 618−624.
Lipmaa, H; Mielikäinen, T; Laur, S
Private Itemset Support Counting
Seventh International Conference on Information and Communications Security, ICICS ’05, 2005:97−111.